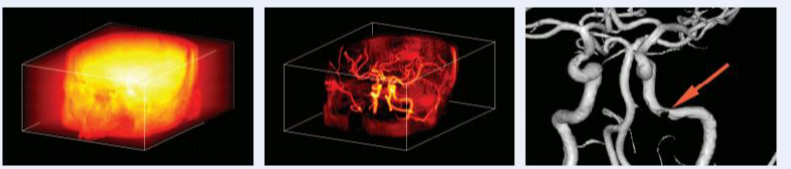
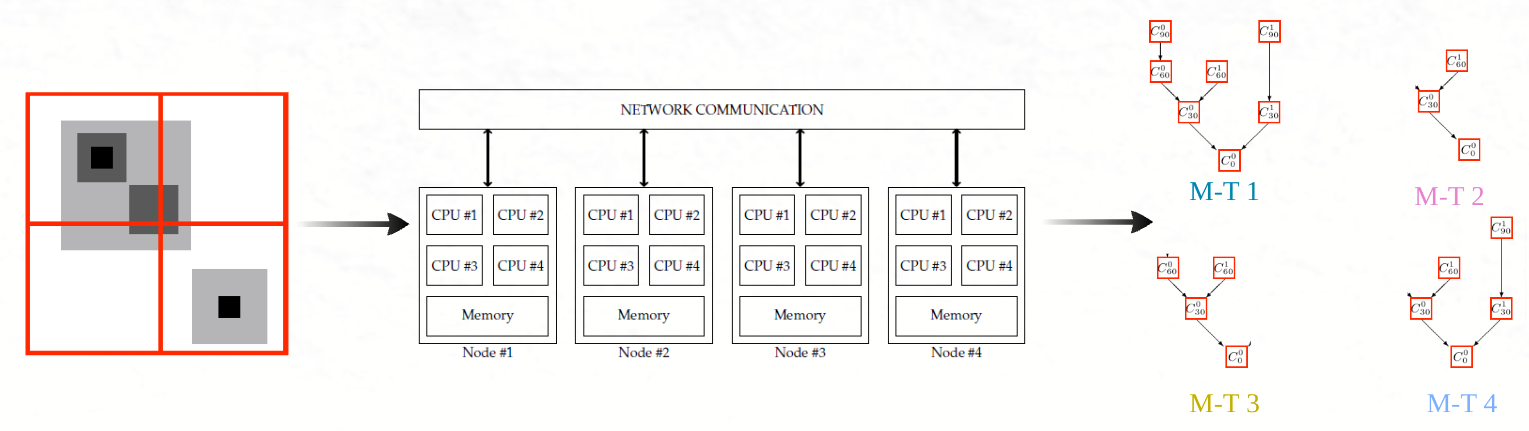
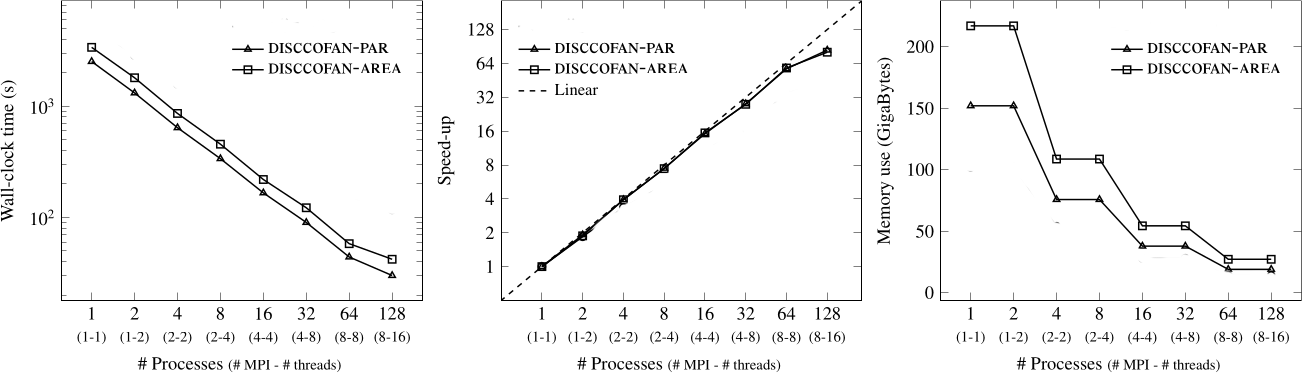
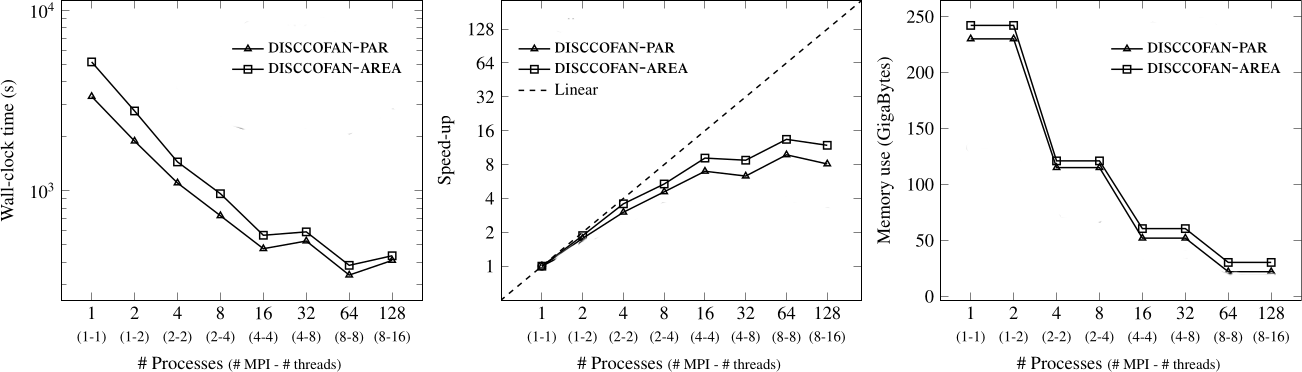
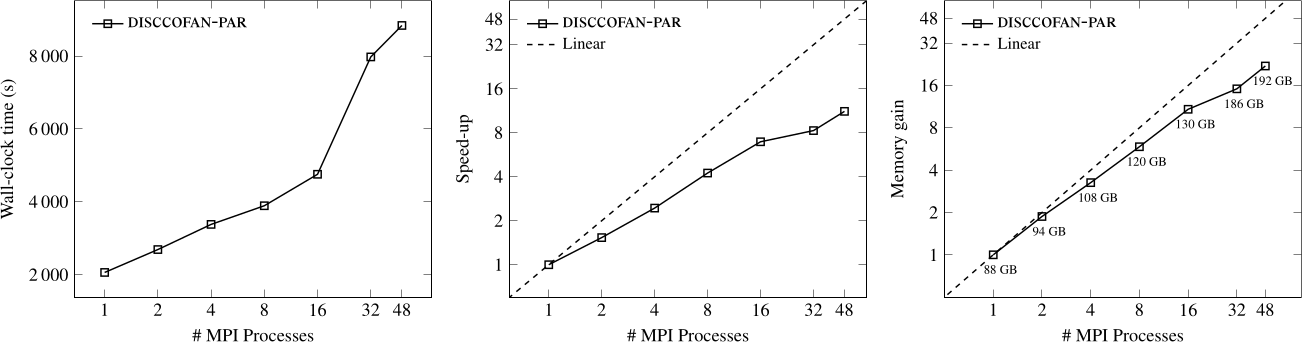
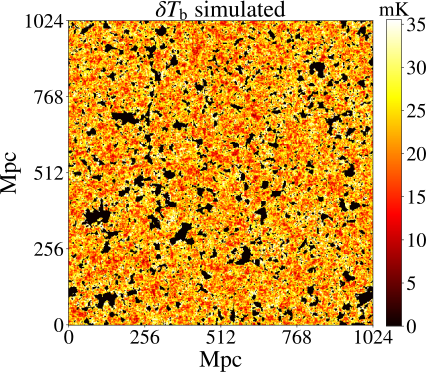
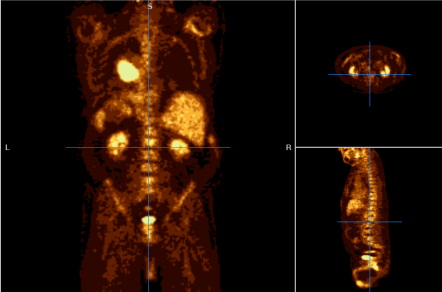
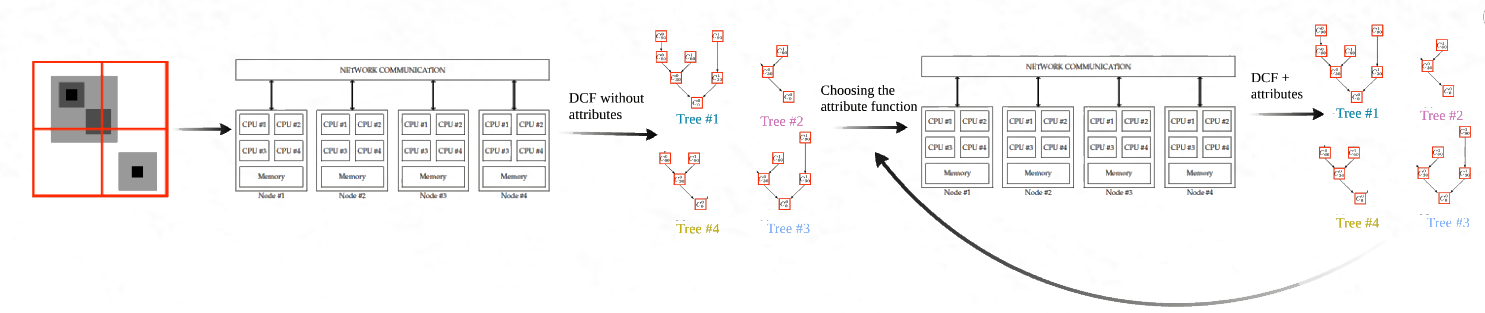
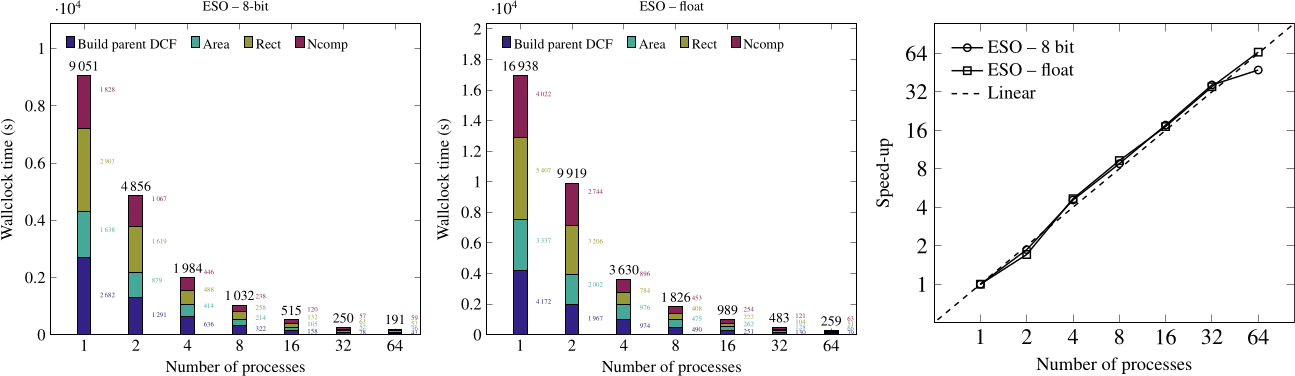Connected operators and Component trees.
Connected operators and component trees are image processing tools used primarily for filtering and simplifying images while preserving important structures. Connected operators act by removing or altering image components based on criteria like size or intensity, without affecting object boundaries. Component trees, on the other hand, are hierarchical structures that represent image regions (or components) at various levels of granularity. They enable efficient analysis and manipulation of the image’s structure, making it easier to apply targeted transformations while retaining meaningful shapes or features in the image. These tools are widely used in areas like computer vision and medical imaging. I have worked on improving certain techniques and deploying a software using them.
Introducing the Connected Component Trees
A component tree works by decomposing an image into regions that are connected by pixels sharing similar intensity levels. The tree is built by progressively merging connected components as the intensity threshold changes, resulting in a hierarchical representation where the root of the tree represents the entire image, and the leaves correspond to the smallest connected components at specific intensity levels.
This hierarchical structure allows for a detailed representation of the image’s content at different levels of abstraction. For example, if you consider a grayscale image, a component tree can be built by analyzing the connected regions of pixels at each grayscale intensity level (or threshold). As the threshold increases, new components emerge or merge with existing ones, creating branches and nodes in the tree. Each node in the tree represents a connected component of the image, and the relationships between nodes (parent-child relationships) capture how regions are connected across different intensity thresholds.
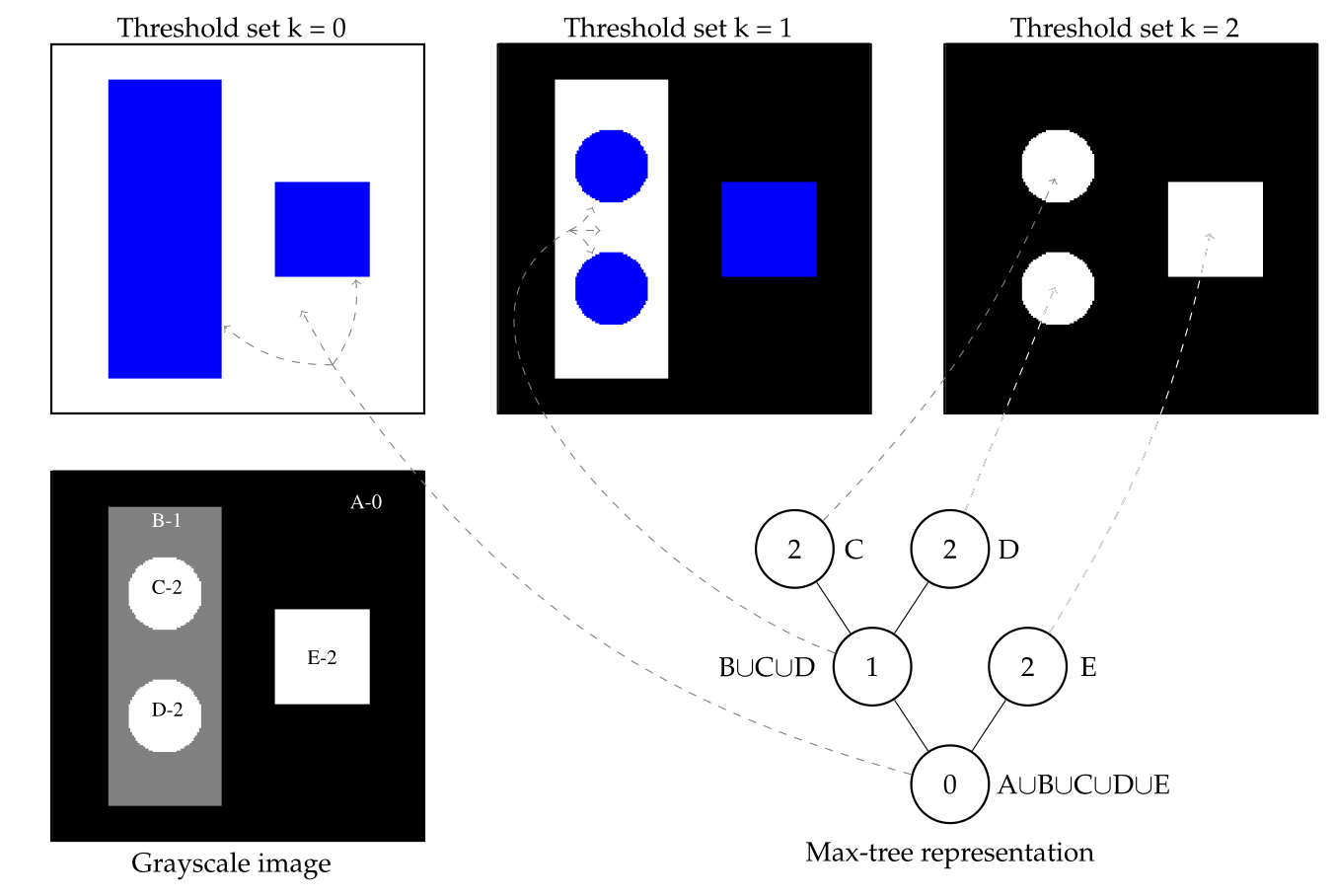
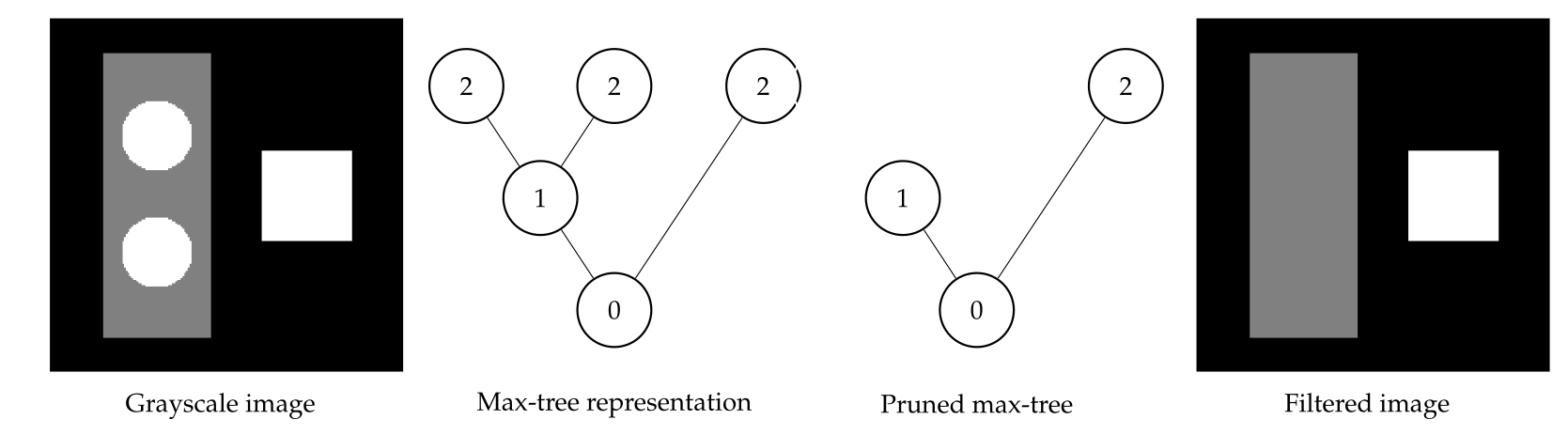
Component trees offer several advantages that make them powerful tools for image analysis. First, they allow for efficient image filtering by manipulating connected components based on their properties, such as size, shape, or intensity. This makes it easy to remove noise, highlight important structures, or simplify the image without distorting object boundaries, which is crucial in fields like medical imaging where object precision is important.
Another fascinating aspect is their ability to represent hierarchical information. Component trees give you a multi-scale view of the image, meaning you can analyze features at different levels of detail, from fine textures to larger regions. This enables adaptive filtering, where you can apply different transformations to various levels of the tree depending on the analysis task. For example, small structures like noise can be easily filtered out, while important large-scale features remain intact. Find some example of applications below