Localizing molecules in super-resolution microscopy
From March to August 2016, as part of the Morpheme Team in Sophia Antipolis, I worked under the supervision of Emmanuel Soubies and Laure Blanc-Féraud to implement a new algorithm to perform high-precision molecule localization using photo-activated localization microscopy techniques (PALM).
Superresolution microscopy is a cutting-edge imaging technique that allows scientists to see very small structures at a much higher resolution than traditional microscopes. Regular microscopes are limited in their ability to resolve details because of the way light behaves. They often produce blurry images when looking at objects that are very close together.
Superresolution microscopy overcomes this limitation by using special methods to achieve a clearer view. It can visualize objects at the nanoscale—much smaller than what conventional microscopes can detect. This means scientists can see details of cells, proteins, and other tiny biological structures with incredible clarity.
By providing sharper images, superresolution microscopy helps researchers understand complex biological processes, develop new medical treatments, and explore the fundamental building blocks of life. It’s a powerful tool that opens up new possibilities in scientific research and innovation.
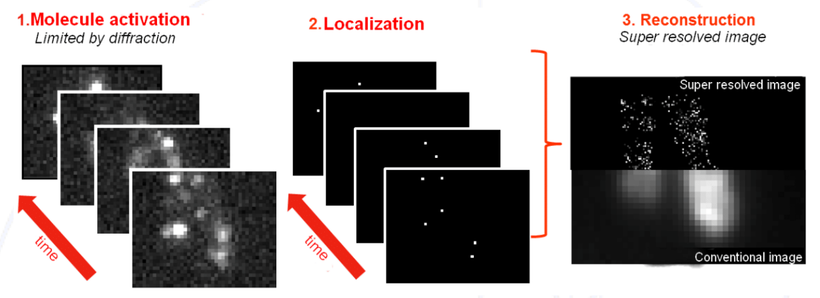
Reconstructing the molecules' position
Developing algorithms to reconstruct the positions of molecules in superresolution microscopy involves several key steps. First, scientists capture many images of fluorescently labeled molecules, which glow when excited by light. Each image shows these molecules as blurry spots. Algorithms analyze these spots to pinpoint the exact locations of the molecules by fitting mathematical models to the observed data.
By combining information from multiple images, the algorithms can create a detailed map of where each molecule is located. This process allows researchers to study the organization and interactions of molecules within cells, leading to new insights in biology and medicine.

In 2016, I worked for 6 months with mathematiciens and biologist to build such an algorithm. We obtained very promising results with a method we later called SMLM-CEL0, and which was published and presented at the ISBI 2017 conference in Australia. On the right, you can check out a small animation showing how an example of live reconstruction, where the positions of each molecules is successively added to the picture to construct the super-resolved images
